Current Release: Release v3.0 : 14th Sep, 2018
We plan to update the Lynx knowledgebase with the latest versions from the sources every 4-Months:
Current DB Updates: v2018.1 : Sep 2018
Lynx aims at providing an informatics platform that will enable the development of efficient, scalable and modular research focused biomedical applications. In order to leverage the enormous amount of biomedical data available and accommodate the exponentially growing data, Lynx provides an infrastructure that systematically integrates the data into a knowledgebase, a modular Java-based enterprise platform to access the integrated data, a schema driven data delivery interface (REST based Web services) and Web2.0 based User Interfaces and visualization to infer the intrinsic topology and unknown patterns.
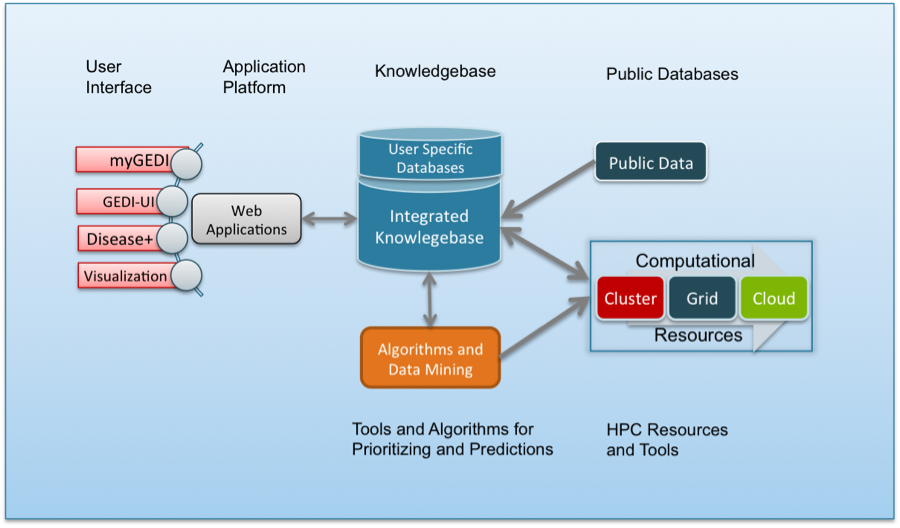
The Lynx Knowledge Base (Lynx KB) integrates information from multiple heterogeneous resources: public databases, text-mining engines, clinical and experimental data provided by the users, results of analyses performed by collaborators via Web-services (e.g. VISTA project) and users annotations. Lynx KB is a relational data warehouse that integrates genomic, pathways, phenotypic, disease independent and disease-specific data and ontologies from over 35 sources.
Type of Data Sources Genomic NCBI, ENSEMBL, UNIGENE, Transfac, VISTA Proteomic BIND, BioGRID, HPRD, MINT, Uniprot, Interpro Pathways – related KEGG, REACTOME, NCI, BIOCARTA, STRING Disease-specific OMIM, Genecards, MESH, Disease Ontology, AUTDB, cancer Gene Index, AGRE, LisDB Phenotypic OMIM, Human Phenotype Ontology Variations Genomic Association Database, Database of Genomic Variants, Human Genomic Mutation Database, SLEP, NHGRI, dbSNP, VISTA Text-Mining Geneways
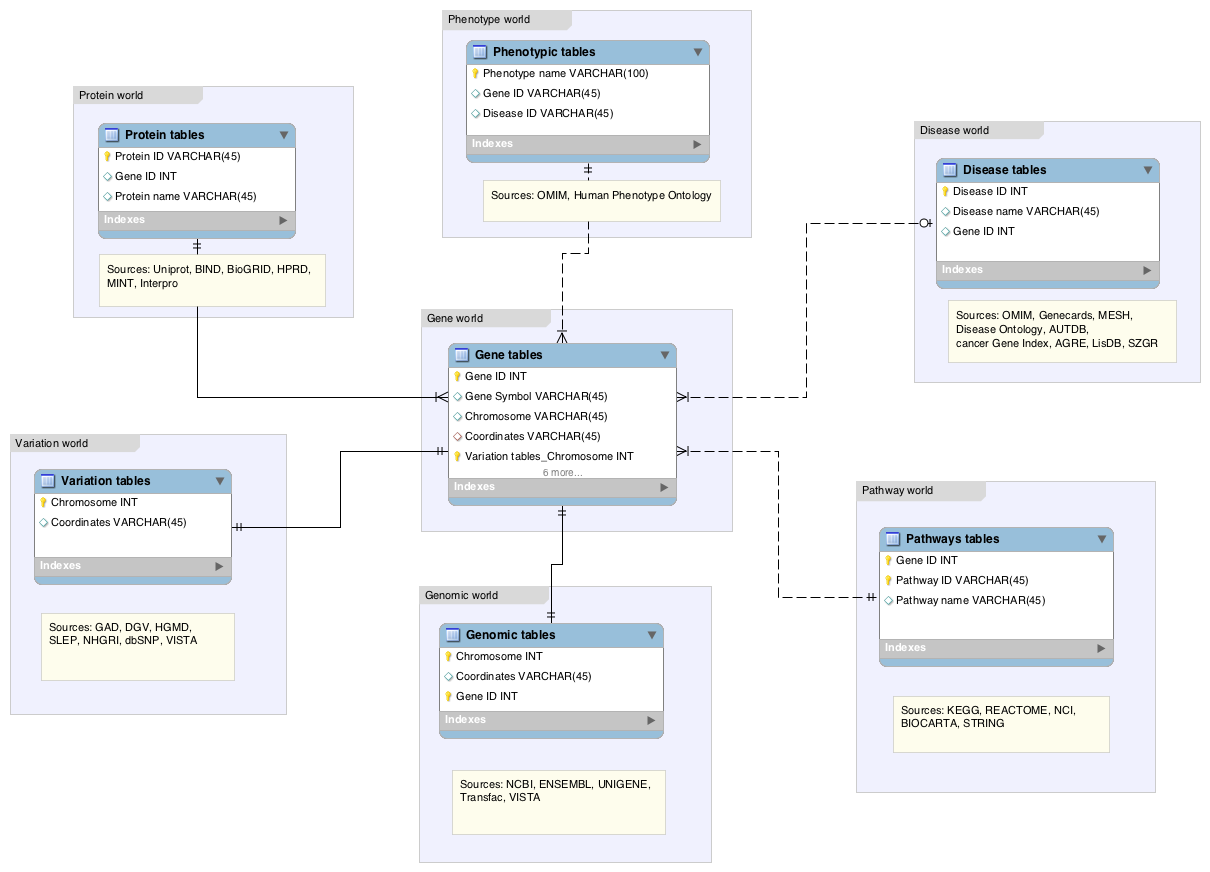
Data in the Lynx KB is modeled, cross-referenced and stored in a relational database. An ER diagram depicting our model is shown below.
Lynx is being developed as a Spring and Java based Enterprise platform based on a scalable, modular and extensible Service-Oriented Architecture (SOA) to support the development of Biomedical applications.
The Lynx's SOA is designed on the principles of ‘Separation of Concern’ and ‘Aspect-Oriented Programming (AOP)’ paradigm. The different tiers are interweaved together using dependency injection using Spring. Figure 2, shows the system architecture with various components, tiers and how they interact with each other.
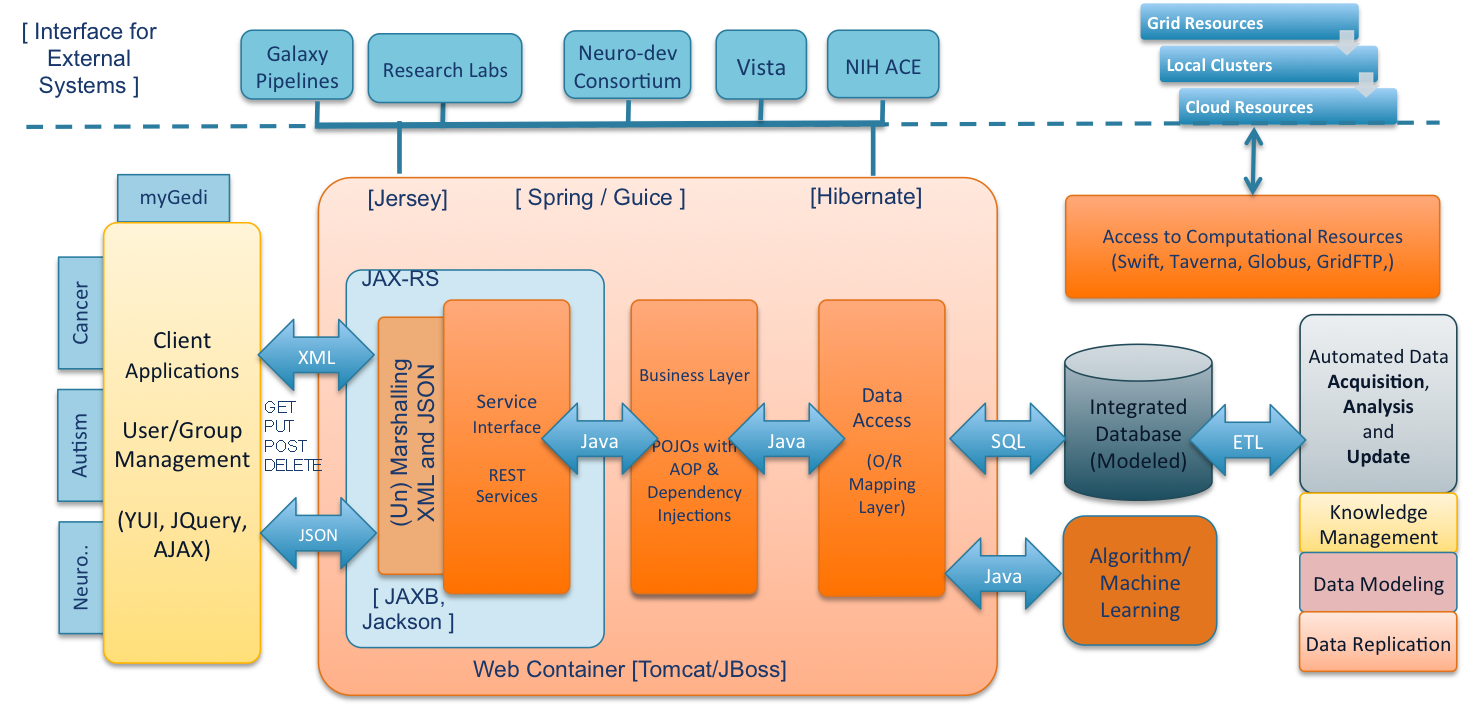
The Service-Oriented Architecture enables Lynx Platform functionalities to be provided as services (RESTful Services) and thus allows developing applications that consume these services. As mentioned in the previous section, these services (domain objects) provide a schema (XSD) and message-based interaction with the applications. Lynx uses Jersey, a reference implementation of JAX-RS for building RESTful Web services. Jersey together with JAXB be automatically handles marshaling and un-marshaling of XML requests and results. For example, the domain specific objects returned by the Domain layer are automatically translated back to XML data as per the definition of XSD and the service layer serves this XML data back to the client. Lynx's service layer also supports JSON formats as return types.